Short Communication 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Diversity of Alkane-1-Monooxygenase Genes in Actinobacteria of the Genus Rhodococcus
*Corresponding author: Puntus Irina Filippovna, G.K. Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, prospect Nauki 5, Pushchino, Moscow Region, Russia.
Received: May 31, 2022; Published:June 09, 2022
DOI: 10.34297/AJBSR.2022.16.002246
Abstract
By analyzing genetic similarity of rhodococci actinobacteria seven phylogenetic groups are allocated on the base on alkane- 1-monooxygenases sequences. R. qingshengii F2-2 genome have been shown to have alkane-1-monooxygenase genes affiliated to five different types. We assume the use of alkane-1-monooxygenase genes diversity as a molecular marker to identify Rhodococcus species.
Keywords: Rhodococcus, Alkane degradation, Alkane-1-monooxygenase genes, classification
Short Communication
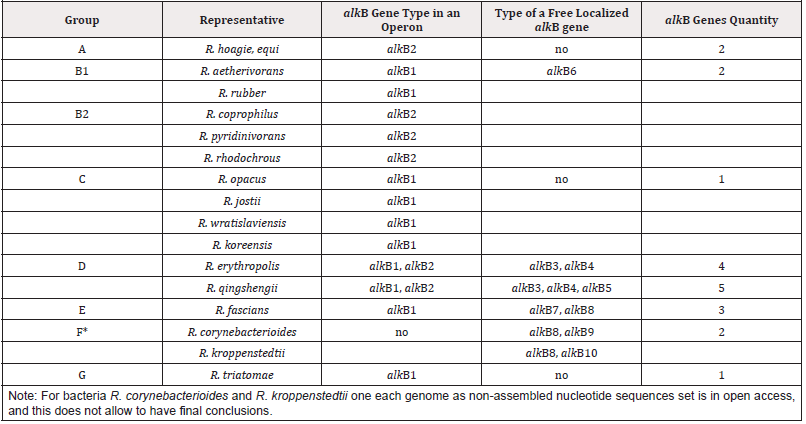
Rhodococci actinobacteria can degrade hydrocarbon compounds representative of various chemical structures and are used in bioremediation technologies. Alkane-1-monooxygenase is the first enzyme catalyzing alkane cleavage, transforming alkanes into primary alcohol. We analyzed protein sequences that include functional domains distinctive for the known rhodococcal alkane- 1-monooxygenases: Hist1 (HELGHK), Hist2 (EHNXGHH), Hist3 (LQRHSDHHA) and HYG (NYLEHYGI). Search for homologous alkane-1-monooxygenases of rhodococci (threshold E value below 1e-156) was performed in an NCBI database using psi-blast utility. Using USEARCH software (version 11.0.667_i86linux32) [1] proteins were excluded whose homology to an alkane-1- monooxygenase did not exceed 60%. Amino acid sequences of homologous alkane-1-monooxygenases were aligned with a help of Clusta lX, version 2.1 [2] and then used to search for functional domains using MEME software, version 5.1.1 [3]. Search for regulatory sequences was performed using Sigmo ID software [4]. We have revealed that the quantity of alkB genes and their genetic organization depend on taxonomic affiliation of bacteria under study (Table 1). Phylogeny of alkane-1-monooxygenase corresponds to phylogeny of Rhodococcus bacteria offered based on comparing genomes, vital genes, conservative proteins, DNA-DNA hybridization, and physiologo-biochemical traits [5]. By analyzing genetic similarity of rhodococci bacteria seven phylogenetic groups are allocated: A, B, C, D, E, F and G, with B (B1 and B2) group and E (E1 and E2) one having the highest polymorphism. Due to Table 1, most typical representative of A group is R. hoagie/R. equi; B1- R. aetherivorans, R. ruber; B2 - R. coprophilus, R. pyridinivorans, R. rhodochrous; C - R. opacus, R. jostii, R. wratislaviensis, R. koreensis; D - R. erythropolis, R. qingshengii; E - R. fascians; G - R. triatomae.
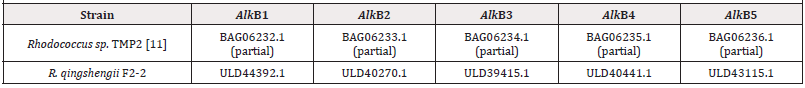
Rhodococcus qinqshengii F2-2 strains with high petroleumutilizing activity was isolated from soils sampled from oil deposit Festivalnoe in Western Siberia, Russia [6]. The strain grows in media with crude oil and diesel fuel, produces tregalolipid biosurfactants and can use alkanes and benzene as a sole source of carbon and energy. In the study the genome of Rhodococcus qinqshengii F2-2 was sequenced using technologies of Oxford Nanopore and Illumina MiSeq and assembled. Genome includes a chromosome of 6.3 mb [7], two linear [8-9] plasmids of 156 (pLP156) and 337 kb (pLP337), and a circular one, pCP209, of 209 kb [10,11]. Surprisingly, it has managed to find a gene cluster encoding for phenazine synthesis on pLP156, that includes five genes: phzF, trans-2,3-dihydro-3- hydroxyanthranilate isomerase (EC:5.3.3.17), phzE, 2-amino- 4-deoxychorismate synthase (EC:2.6.1.86), phzA_B, phenazine biosynthesis protein, phzG, dihydrophenazinedicarboxylate synthase (EC:1.10.3.16), and phzS, 5-methylphenazine-1- carboxylate 1-monooxygenase (EC:1.14.13.218). Genome has 6741 coding sequences (CDS), three rRNA clusters (5S, 16S and 23S) and 59 tRNAs. 5234 CDS can be affiliated to 25 different clusters of orthologous genes. R. qingshengii F2-2 genome has five alkane-1- monooxygenase genes (Table 2).
The gene alkB1 has a classic organization, namely, is in an operon presented by five genes (alkB1, rubA1, rubA2, rubB and alkU1). Three genes of the operon being electron carriers (rubredoxins encoded by rubA1 and rubA2 and a rubredoxin reductase encoded by rubB) provide an alkane-1-monooxygenases functional activity. The gene alkB2 is part of operons (alkB2, rubA3, rubA4 and alkU2) with the last one’s non-possessing determinants encoding for rubredoxin reductases. Thus, in genomes of rhodococci two type of alkB genes have managed to distinguish that are part of differently arranged operons, encoding alkane-1-monooxygenases of two types-AlkB1 и AlkB2, correspondingly. Herewith AlkB1 type enzymes are typical for non-pathogenic degrader bacteria affiliated to B1 phylogenetic group (R. aetherivorans, R. ruber), C (R. opacus, R. jostii, R. wratislaviensis, R. koreensis) and G (R. triatomae) and plant pathogens of E group (R. fascians). Enzymes of AlkB2 type are synthesized in cells of B2 group-affiliated degrader bacteria (R. coprophilus, R. pyridinivorans, R. rhodochrous) and A group animal pathogens (R. hoagie/equi). D group is on a special place that includes degrader bacteria and best studied biosurfactant producers (R. erythropolis, R. qingshengii) that synthesize two types of phylogenetically distant alkane-1-monooxygenases. AlkB1 type is the closest to the same one in G group bacteria (R. triatomae), and AlkB2 type demonstrates the greatest similarity to the same one of A group pathogenic bacteria (R. hoagie/equi). The highest quantity of distinct alkB genes (alkB3, alkB4, alkB5) is detected in bacterial genomes belonging to D phylogenetic group (R. erythropolis, R. qingshengii). Herewith, genes alkB3 and alkB4 are present in all the representatives of this group, whereas alkB5 genes are detected in R. qingshengii strains only [12].
Based on the data analysis performed it may be concluded that phylogenetically most diverse alkane-1-monooxygenases are produced by R. erythropolis, R. qingshengii belonging to group D (from four to five types of the enzymes). Separately localized alkB determinants would seem to emerge owing to duplication of alkB genes occurring in the corresponding operons, and further divergence process of gene copies proceeds. However, their conservative localization contradicts the last assumption: once appeared, alkB genes keep their localization, only their nucleotide sequences are changing. This situation is possible only because of horizontal gene transfer, much common process in prokaryotes. Understanding the genome formation of bacteria important for human is essential since cognate proteins must realize similar functions. Alkane-1-monooxygenase genes appear to be a molecular marker to identify Rhodococcus species. The patterns revealed in this study complement the available to date data and can be useful to determine taxonomy and phylogeny of closely related rhodococci.
Acknowledgments
The reported study was funded by RFBR* and BRFBR**, project number 20-54-00002 “Peculiarities of the synthesis of surfaceactive compounds by bacteria that efficiently utilize oil at low and high temperatures”.
*Russian Foundation for Basic Research; **Belarusian Foundation for Basic Research.
Upon admission, initial laboratory tests demonstrated: FBC: WBC 9690/μL, Neutrophil-Lymphocyte Ratio (NLR) 8, indicating mild stress level, Haemoglobin 9.7gr/dL, Haematocrit 33.3%. The patient’s biochemical profile was as follows: glucose 228 mg/dL , urea 53 mg/dL , creatinine 0.86 mg/dL, Κ 4.1mmol/L, Na139 mmol /L , SGOT 10U/L, SGPT 13U/L , γGT 11U/L, ALP 37U/L, amylase 27.1 U/L, CRP 0.50mg/dL.
Further sample testing included urine chemical analysis using an automated urine analyzer (iChem Velocity Urinalysis Analyzer, Leriva), urine sediment microscopy and conventional urine culture. Urine microscopy showed pyuria, mild hematuria and the presence of cocci. After 48-hour incubation, the urine culture revealed 10^5cfu/mL of a Gram (+) positive, catalase (-) negative, alpha-hemolytic coccus in tetrads, on blood agar. The identification testing was performed using the semi-automated Microscan Autoscan-4 System (Siemens). The microorganism was identified as Aerococcus urinae (Microscan ID: 99.99%) and was susceptible to penicillin, ampicillin, ciprofloxacin, levofloxacin and vancomycin, and resistant to nitrofurantoin, according to the criteria of the European Committee on Antimicrobial Susceptibility Testing (EUCAST).
Conflict of Interest
The authors declare no conflict of interest.
References
- Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19): 2460-2461.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21): 2947-2948.
- http://meme-suite.org/index
- Nikolaichik Y, Damienikan AU (2016) SigmoID: a user-friendly tool for improving bacterial genome annotation through analysis of transcription control signals. PeerJ 4: e2056.
- Sangal V, Goodfellow M, Jones AL, Schwalbe EC, Blom J, et al. (2016) Next-generation systematics: An innovative approach to resolve the structure of complex prokaryotic taxa. Sci Rep 6:1-12.
- Puntus IF, Borzova OV, Funtikova TV, Suzina NE, Egozarian NS, et al. (2019) Contribution of soil bacteria isolated from different regions into crude oil and oil product degradation. J Soils Sedim 19: 3166-3177.
- Rhodococcus qingshengii strain F2-2 chromosome, complete genome.
- Rhodococcus qingshengii strain F2-2 plasmid pCP209, complete sequence.
- Rhodococcus qingshengii strain F2-2 plasmid pLP337.
- Rhodococcus qingshengii strain F2-2 plasmid pLP156.
- Takei D, Washio K, Morikawa M (2008) Identification of alkane hydroxylase genes in Rhodococcus strain TMP2 that degrades a branched alkane. Biotechnol Lett 30(8): 1447-1452.
- Ratnikova MS, Titok MA (2020) Molecular genetic markers for identification of Rhodococcus erythropolis and Rhodococcus qingshengi. Mikrobiologiya 89(4):435-442.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.